Data classification using Neural Networks and Genetic Programming
01 July 2018
Plamen Kolev
Genetic Programming
Revised justification
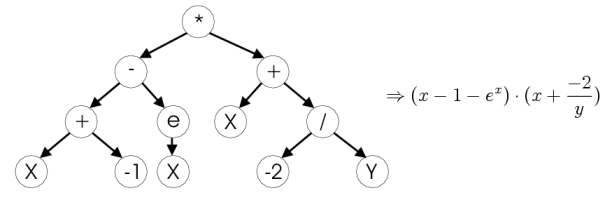 As the first biologically inspired machine learning algorithm, I decided
to use Genetic Programming. Genetic Programming is suitable for
classifying multiple classes. An Oxford journal about classifying
microarray datasets talks about ways GP can be used to classify and
diagnose particular types of cancer (Kun-Hong L. et al. 2008). An
implementation of a Genetic Programming for classification works by
applying divide and conquer strategies to the training data, splitting
it into multiple sets of decision trees. The leaves of the trees
represent different classes (Jeroen Eggermont et al. 2004).
As the first biologically inspired machine learning algorithm, I decided
to use Genetic Programming. Genetic Programming is suitable for
classifying multiple classes. An Oxford journal about classifying
microarray datasets talks about ways GP can be used to classify and
diagnose particular types of cancer (Kun-Hong L. et al. 2008). An
implementation of a Genetic Programming for classification works by
applying divide and conquer strategies to the training data, splitting
it into multiple sets of decision trees. The leaves of the trees
represent different classes (Jeroen Eggermont et al. 2004).
In the original proposal, I make the case for using Genetic Algorithm with k-nearest neighbours, but later decided to not use it, as GP frameworks such as epochx contain all the necessary libraries and tuning strategies that will provide a better result with the ability to do more fine-grain tuning. It will also allow me to explore a more generic solution.
In genetic programming, each decision branch is formed by a mathematical expression (functions). For the dataset, I experimented with different ‘syntax’ functions. The findings are documented in the tuning section for this algorithm, but the most effective were the trigonometric ones.
Description of implementation
As mentioned above, I used the epochx library framework to bootstrap
my implementation. I created a new source package called GP, which
contains GPClassifier.java, GPControl.java and GPWrapper.java. The
implementation of the GPControl class is very similar to the provided
sample one, as its purpose is to test the actual implementation. Minor
modifications were made to accommodate for the newly created data
structures. Another modification is exposing the generateSubSolution
to return GPClassifier, as this class is used in the main method to
write performance and configuration statistics to a report file called
gpstats.csv.
The GPClassifier class contains a genetic program instance that has
learned on the training data and implements classifyInstance and
printClassifier. Those methods are passed to the underlying genetic
programming class GPWrapper, the class is acting as a middle man
between the control and the genetic programming class.
The GPWrapper class contains all the necessary constructs and tunable
variables for the epoch framework,which are described in the tuning
section.
GPWrapper.java constructor.
``` java syntax.add(new SignumFunction()); syntax.add(new SineFunction()); syntax.add(new CosineFunction()); syntax.add(new ArcTangentFunction()); syntax.add(new AbsoluteFunction());
syntax.add(new AddFunction()); syntax.add(new DivisionProtectedFunction()); syntax.add(new SubtractFunction()); … for (int i = 0; i < Attributes.getNumAttributes(); i++) { syntax.add( variables[i] = new Variable(“dim” + i, Double.class) ); } ```
The constructor allows for an arbitrary attributes to be passed as the syntax, but the syntax functions are specially tuned for the given dataset, as trigonometric functions are suitable for classifying the circular representation of the data.
An auxiliary function called parseData is provided just to store the
data in an internal array for later usage.
This class also returns itself as an instance when generateClassifier
is called. The method is responsible for setting the different tuning
parameters and training the genetic program.
GPWrapper.java classifyInstance method.
java
public int classifyInstance(Instance ins){
GPCandidateProgram a = (GPCandidateProgram) this.getProgramSelector().getProgram();
for (int i = 0; i < Attributes.getNumAttributes(); i++) {
this.variables[i].setValue(ins.getRealAttribute(i));
}
Double result = (Double) a.evaluate();
return (int) Math.round(result);
}
The classifyInstance method is used after the GP is trained to
determine the class of the data (black or white).It uses the
GPCandidateProgram object with the best fitness (closest to 0). That
program is given the instance attributes and evaluates a solution. The
output is an integer that represents the class.
As the GPWrapper extends from the epochx’s GPModel, it also
implements getfitness method. This method is applied to each member of
the population (candidate program). Each candidate produces an
output when given the input data points from the training set. If the
output of the program matches the class (predicts it), a score variable
is incremented. The best candidate program should have a score which
should be as close to the size of the training set as possible.
printClassifier simply finds the fittest program from the candidate
pool and prints its representation, which is just a string of nested
functions.
Similar to the NeuralClassifier, this class also contains setconfig
function, which allows for environmental variables to be set and used to
tune different parameters. Finally, the classifier also implements
visualise, which draws a representation of the training and test data to
the screen using Jpanel in a class called Picasso.
Tuning for the provided dataset
For the tuning, I exposed a variety of parameters that are used when creating and training the classifier. The most important tunable parameter is the population size, which represents the number of generated random program candidates. A large population range between 600 - 1200 members provided enough diversity to generate the most efficient classifiers and below 600, the accuracy relied mainly on the random seed value and luck, making the predictions unreliable.
I also decided to experiment with the generation size. When a large generation set is configured, at some point the population diversity evens out and the fittest individual does not change much. A value between 20-70 generations is the most appropriate, as it gives the programs enough time to evolve and mutate, but keeps the running time short.
Another important factor in the efficiency was selecting the syntax
functions when creating the program trees. They had very significant
impact on the evaluation time and efficiency.The performance of each
function varied, as some were very beneficial, some had not much impact
and others had a negative effect . The implementation does not include
flags for tuning which type of function to use, but rather includes a
selection of the the most effective ones. The highest result I was able
to achieve is using SignumFunction as well as the trigonometry
functions. I included division, subtraction and addition, as they helped
with the program’s diversity.
Experimenting with number of selected elite members, crossover, mutation and reproduction probability did not provide a significant difference during my experiments.
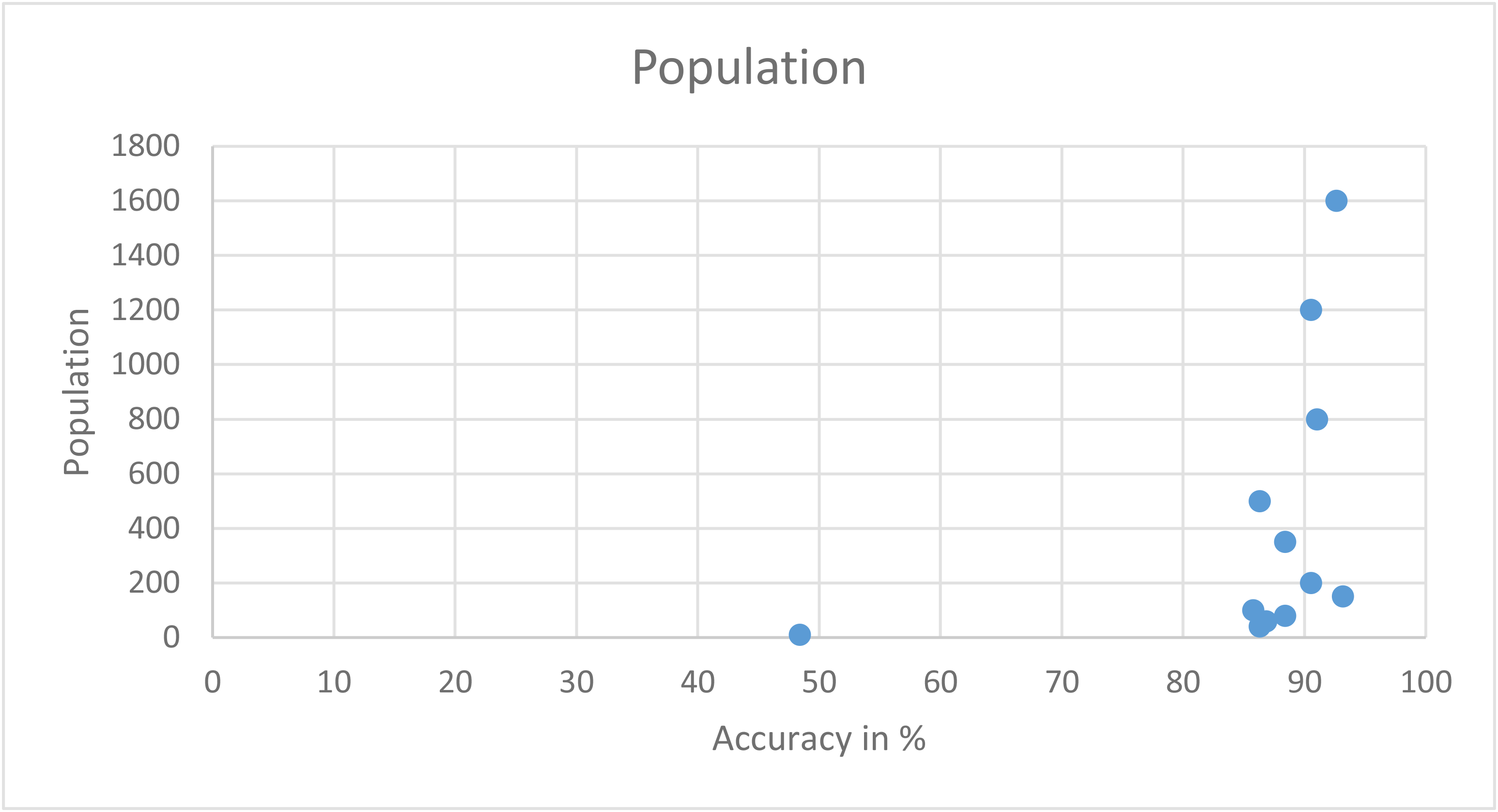
Performance report
| Runtime | Efficiency in % | Population size |
|---|---|---|
| 0.506 | 48.421052631578945 | 10 |
| 7.528 | 86.31578947368422 | 40 |
| 10.396 | 88.42105263157895 | 80 |
| 30.763 | 90.52631578947368 | 200 |
| 53.695 | 86.31578947368422 | 500 |
| 163.121 | 90.52631578947368 | 1200 |
| 232.42 | 92.63157894736842 | 1600 |
GP algorithm with different population size
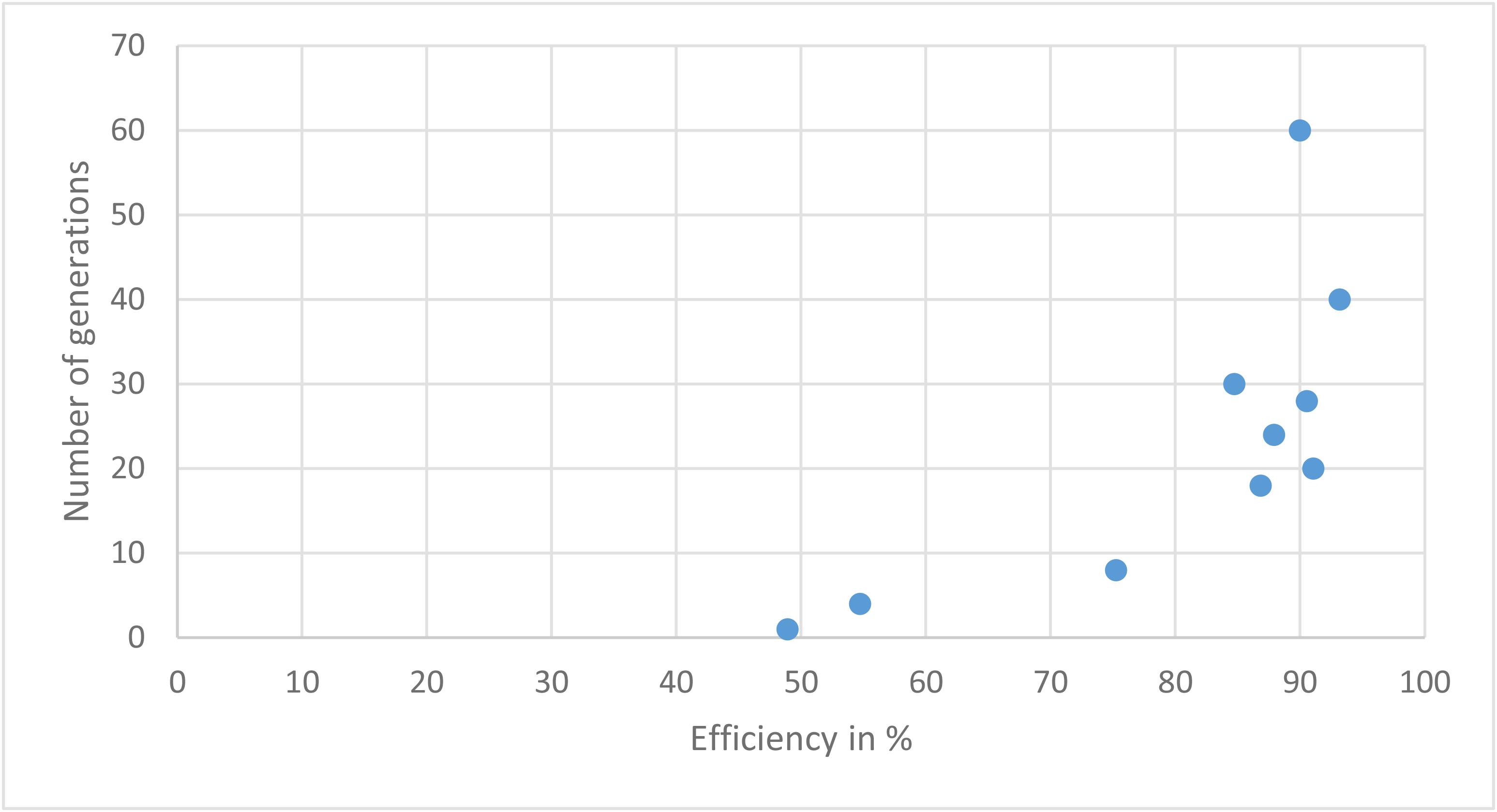
| Time | Efficiency in % | Generation number |
|---|---|---|
| 1.087 | 48.91304347826087 | 1 |
| 1.267 | 54.736842105263165 | 4 |
| 2.045 | 75.26315789473685 | 8 |
| 3.582 | 86.8421052631579 | 18 |
| 12.429 | 91.05263157894737 | 20 |
| 10.704 | 87.89473684210526 | 24 |
| 17.317 | 90.52631578947368 | 28 |
| 16.921 | 84.73684210526315 | 30 |
| 14.252 | 93.15789473684211 | 40 |
| 25.816 | 90.0 | 60 |
Table showing runs with different generations (stopping criteria)
I also experimented with different crossover probability, number of elite chromosomes and mutation but they did not improve or lower the efficiency. The difference I noticed is the runtime, as these parameters allow the GP to perform additional operations pore often. I have included a table with my findings for completeness.
| Time | Efficiency % | Elite population count |
| 22.11 | 92.63157894736842 | 1 |
| 38.558 | 93.15789473684211 | 4 |
| 42.268 | 90.0 | 12 |
| 34.73 | 97.89473684210527 | 18 |
| 43.304 | 88.94736842105263 | 24 |
| 35.337 | 96.3157894736842 | 30 |
Table of different size of the elite population
| Time | Efficiency in % | Crossover probability |
|---|---|---|
| 77.397 | 90.0 | 0.1 |
| 87.546 | 94.21052631578948 | 0.3 |
| 82.201 | 87.36842105263159 | 0.4 |
| 58.289 | 93.15789473684211 | 0.8 |
| 62.03 | 94.21052631578948 | 1.0 |
Table of different crossover probability
| Time | Efficiency in % | Mutation probability |
|---|---|---|
| 15.015 | 85.78947368421052 | 0.1 |
| 94.777 | 89.47368421052632 | 0.2 |
| 79.374 | 88.94736842105263 | 0.4 |
| 105.695 | 93.6842105263158 | 0.6 |
| 62.391 | 88.42105263157895 | 0.8 |
| 71.862 | 91.57894736842105 | 1.0 |
Mutation probability
Neural Network
Revised Justification
As mentioned in the coursework proposal, I chose Neural network as the method for classification for several reasons: It is suitable for classifying multiple ‘features’ as neural networks have been widely used for speech recognition, facial recognition and malignant cancer detection (Álvarez L. et al. 2009). The Universal approximation theorem also states that a neural network has the ability to can compute every function. Yoshua Bengio and Yann LeCun also mention in their paper that neural networks do not have limitations in the efficiency of the representation of certain types of functions as compared to other approaches.
I also wanted to use a neural network, as it is a very exciting and hot topic in computing and wanted to experiment with the different activation functions and parameters.
Implementation description
Implementing the solution was a multi-part process. During my research, I found it suitable to use Neuroph java framework for the supervised neural network. I then outlined key features and functions that need to be implemented, tuned and integrated. The following is a detailed description of them.
Firstly, I created a special package called Neural, which contains the
main class Control.java of the application and the
NeuralClassifier.java class witch is a wrapper for Neuroph. For the
code of Control.java, I used most of the sample code provided (time
keeping, printing training and test data, etc.) with some minor
modifications. I also replaced the Wrapper classifier with the neural
implementation file NeuralClassifier.java. The new main class was also
modified to allow for stats dumps to csv file used to generate
statistics for the report.
My NeuralClassifier.java extends from the Classifier class and thus
implements classifyInstance and printClassifier. Its purpose is to
be a middle man between the given framework and Neuroph. Because Neuroph
uses its own data representation for the training and testing sets, an
auxiliary function is provided as part of the implementation called
instanceSetToDataSet which parses the InstanceSet and converts it to
DataSet used by Neuroph.
NeuralClassifier.java function instanceSetToDataSet.
java
public DataSet instanceSetToDataSet(InstanceSet trainingData){
int dataRows = Attributes.getNumAttributes();
DataSet td = new DataSet(dataRows, 1);
...
// Data normalisation code skipped
// Data scaling code skipped
td.addRow(new DataSetRow(inputs, new double[]{instance.getClassValue()}));
}
...
}
Here, the TrainingSet is converted to Data set with dynamic rows. This
function is used in the constructor to bootstrap the
MultiLayerPerceptron.
Constructing the actual class is done in the following way:
NeuralClassifier.java function constructor.
java
NeuralClassifier(InstanceSet trainingSet){
...
this.initConfig();
DataSet trainingData = this.instanceSetToDataSet(trainingSet);
this.mlp = new MultiLayerPerceptron(ACTIVATION_FUNCTION,
Attributes.getNumAttributes(), HIDDENNODES, 1);
BackPropagation b = (BackPropagation) mlp.getLearningRule();
b.setMaxIterations(ITERATIONS);
b.setMaxError(MAXERROR);
b.setLearningRate(LEARNINGRATE);
...
this.mlp.learn(trainingData);
}
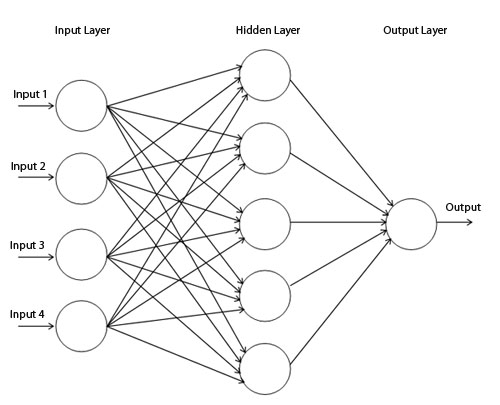
As mentioned in the proposal, I am using Multilayer perceptron with
back-propagation, the above function creates and configures the neural
network instance. As described in the lecture notes, the
MultilayerPerceptron uses an activation function which is one of the
parameters for tuning, I have set the Sigmoid Transfer function as the
default as the highest results are often achieved using it, but others
are explored as part of the performance report. For the input layer, I
use as many neurons as data attributes in the training set, allowing for
a generic solution when applying different datasets.
Neuroph has a construct that allows for the tuning of extra parameters,
like the number of iterations, max errors and learning rate, they are
also configured and demonstrated in the performance section. The given
Attributes, InstanceSet and Instance classes were used throughout,
as they provided helpful methods give the necessary information to make
the solution generic. When going though the source code, I also noticed
that some examples were scaling and normalising the data, so I decided
to include these features into the implementation. I used
DecimalScaleNormalizer.java function normalizeScale example from the
source directory of neuroph to optionally scale down each data point by
a factor of 10 based on a flag. This function and the flag normalise
in instanceSetToDataSet function are extra tunable parameters that I
experimented with but found to cause harm to both the performance and
the efficiency.
NeuralClassifier.java function classifyInstance.
``` java public int classifyInstance(Instance ins) { // provide the classifier with the inputs int numattrs = Attributes.getNumAttributes(); double[] attributes = new double[numattrs]; for (int i = 0; i < numattrs; i++) { attributes[i] = ins.getRealAttribute(i); } this.mlp.setInput(attributes);
// calculate neural network output
this.mlp.calculate();
double[] result = this.mlp.getOutput();
int prediction = (int) Math.round(result[0]);
if(prediction > Attributes.getNumAttributes() || prediction < 0){
return -1;
}
return prediction; } ```
classifyInstance function is used to return a prediction after the
network has gone through the learning process. In this case, the
MultiLayerPerceptron is given the instance data , after which the
probability for each class is computed and returned. The prediction is 0
for black, 1 for white and -1 if it goes outside the domain range, or
more generally, between 0 to n-1 where n is the class range.
NeuralClassifier.java function printClassifier.
java
public void printClassifier() {
for (Double weight : mlp.getWeights()) {
System.out.print("Output weights: " + weight);
}
System.out.println();
}
For printing the final classifier, I grab the weight of each node in the neural network and print it out.
Finally, I created a helper function called initConfig() to read
environment variables that allow for dynamic tuning and shell execution
of the solution for generating statistics. I have included the shell
scripts used for generating the performance data under “gp_shell” for
the genetic programming algorithm and “neural_shell” for the neural
network algorithm. They have been developed and tested under Ubuntu
16.04 and work by using environmental variables. When running the
program, it will create a report file in the current directory with the
performance, duration and the tuned parameters.
Finally, the classifier also implements visualise, which draws a
representation of the training and test data to the screen using
Jpanel in a class called Picasso.
# Description of adjustment and tuning
A neural network allows for a variety of parameter tweaking and
different optimisations.
Reading up on different approaches for training a neural network, The
report (Moriera, M 1995) describes the purpose of a learning rate. The
paper suggests to use a learning rate that is sufficiently large to
allow for the learning process but small enough to guarantee
effectiveness. For that purpose, I used the learning rule function
setLearningRate(value) and found out that a very small value in the
range of 0.01-0.1 usually yields the best results. I experimented with
more than one hidden layers, but often the result was identical or
slightly worse but it increased the learning time, sometimes by a factor
of two. Hence I decided to ignore the tuning of multiple layers, and
instead allow for the tuning of different hidden nodes in the single
layer. Just one layer is used, because the data to classify is not
complex enough to benefit from it (not many features). Tuning the
learning rate had a significant impact on the accuracy and I found that
a lower learning late had a very positive impact on the correctness of
the classifier for a very small performance cost.
Neuroph also provides couple of activation functions as part of the
MultilayerPerceptron package. Experimenting with them, I thought
initially that the gaussian function will yield the best result, but by
far the most efficient function was sigmoid, which consistently
achieves results above 90% accuracy. To contrast that with the tanH
function, it has an extremely negative effect on the accuracy, making
the prediction absolutely wrong. Another function that performed very
poorly is the linear function, which yields 50% accuracy, making the
classifier more similar to a coin toss.
An article on standardising data for neural networks (McCaffrey J. 2014) an argument for normalisation of the data is made. I have allowed for a normalisation flag in my implementation, but enabling it has a negative effect due to the data being evenly distributed and relatively short. As mentioned above, data can also be scaled, but again, this also has a negative effect on the accuracy.
Performance report
| Time in seconds | Efficiency in % | Number of iterations |
|---|---|---|
| 0.189 | 85.26315789473684 | 1 |
| 0.489 | 86.8421052631579 | 20 |
| 0.838 | 87.36842105263159 | 40 |
| 1.121 | 87.89473684210526 | 60 |
| 1.371 | 88.42105263157895 | 80 |
| 1.642 | 88.94736842105263 | 100 |
| 10.2 | 96.3157894736842 | 748 |
| 13.524 | 96.3157894736842 | 997 |
| 26.391 | 97.36842105263158 | 1993 |
| 28.076 | 97.36842105263158 | 2242 |
| 21.338 | 96.84210526315789 | 2989 |
| 32.731 | 96.84210526315789 | 3238 |
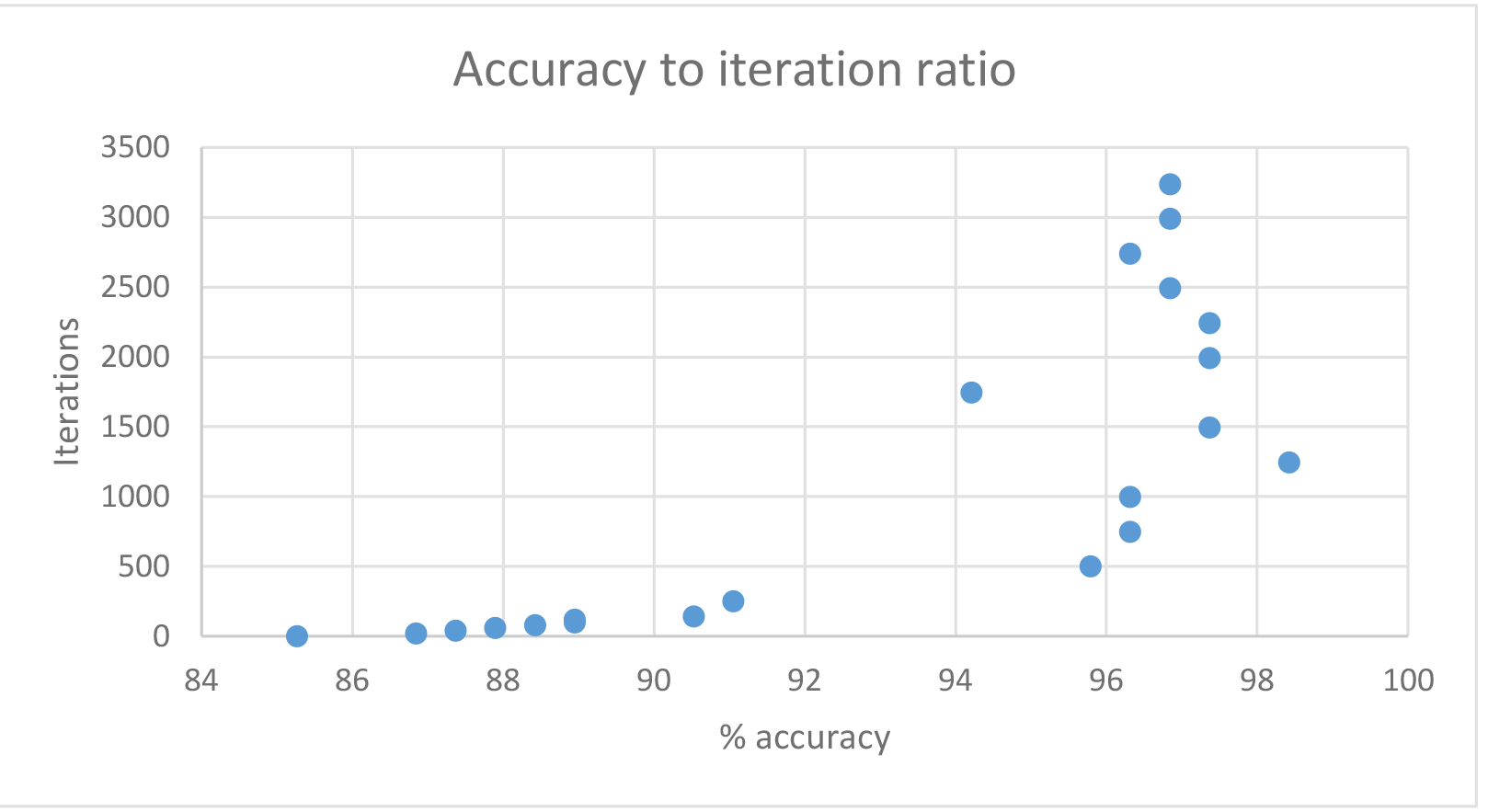
Efficiency with different iterations
The table above shows improvement in performance with the increase of iterations. In my case, it peaked at about 1500-2000 iterations, at which point it did not have a significant impact on the efficiency but it impacted the running time.
| Time | Accuracy in % | Hidden nodes |
|---|---|---|
| 6.379 | 82.10526315789474 | 1 |
| 9.108 | 84.21052631578947 | 3 |
| 12.434 | 91.05263157894737 | 6 |
| 12.125 | 94.21052631578948 | 10 |
| 14.309 | 98.42105263157895 | 17 |
| 16.069 | 97.36842105263158 | 23 |
| 15.644 | 98.42105263157895 | 24 |
| 17.264 | 95.78947368421052 | 29 |
| 17.871 | 97.89473684210527 | 33 |
| 18.304 | 97.36842105263158 | 36 |
| 20.96 | 97.36842105263158 | 40 |
Impact on different hidden nodes
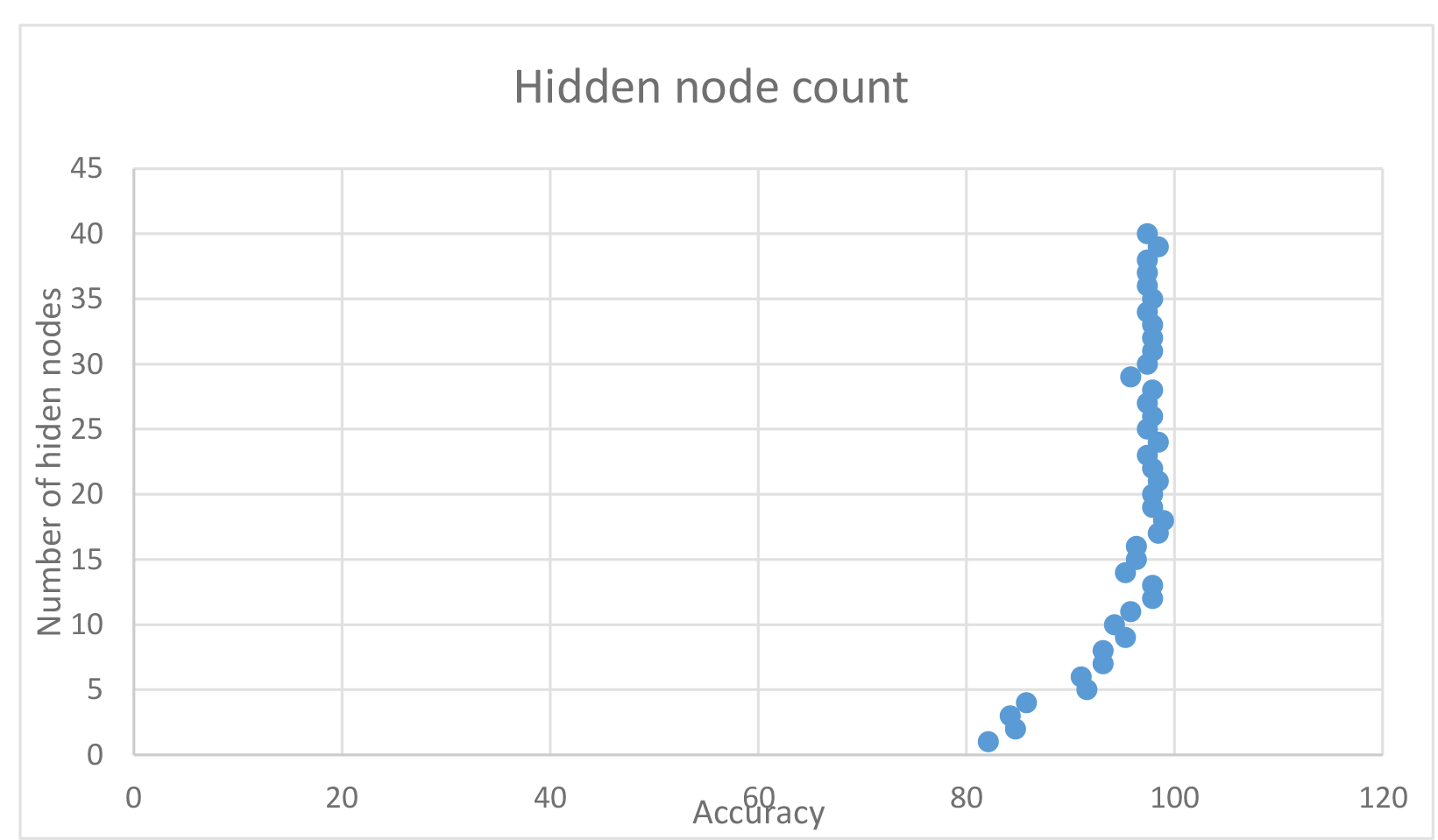
In this case, increasing the amount of hidden nodes had a significant impact on the accuracy, with it peaking at about 17-24 nodes. I found that using 18 hidden nodes fairly reliably produced around 98% accuracy, thus chose it as the base for some runs.
To keep this report short, I will include a graph of learning rate and max error results without including the tables, but the data can be found and viewed under neuralstats.csv
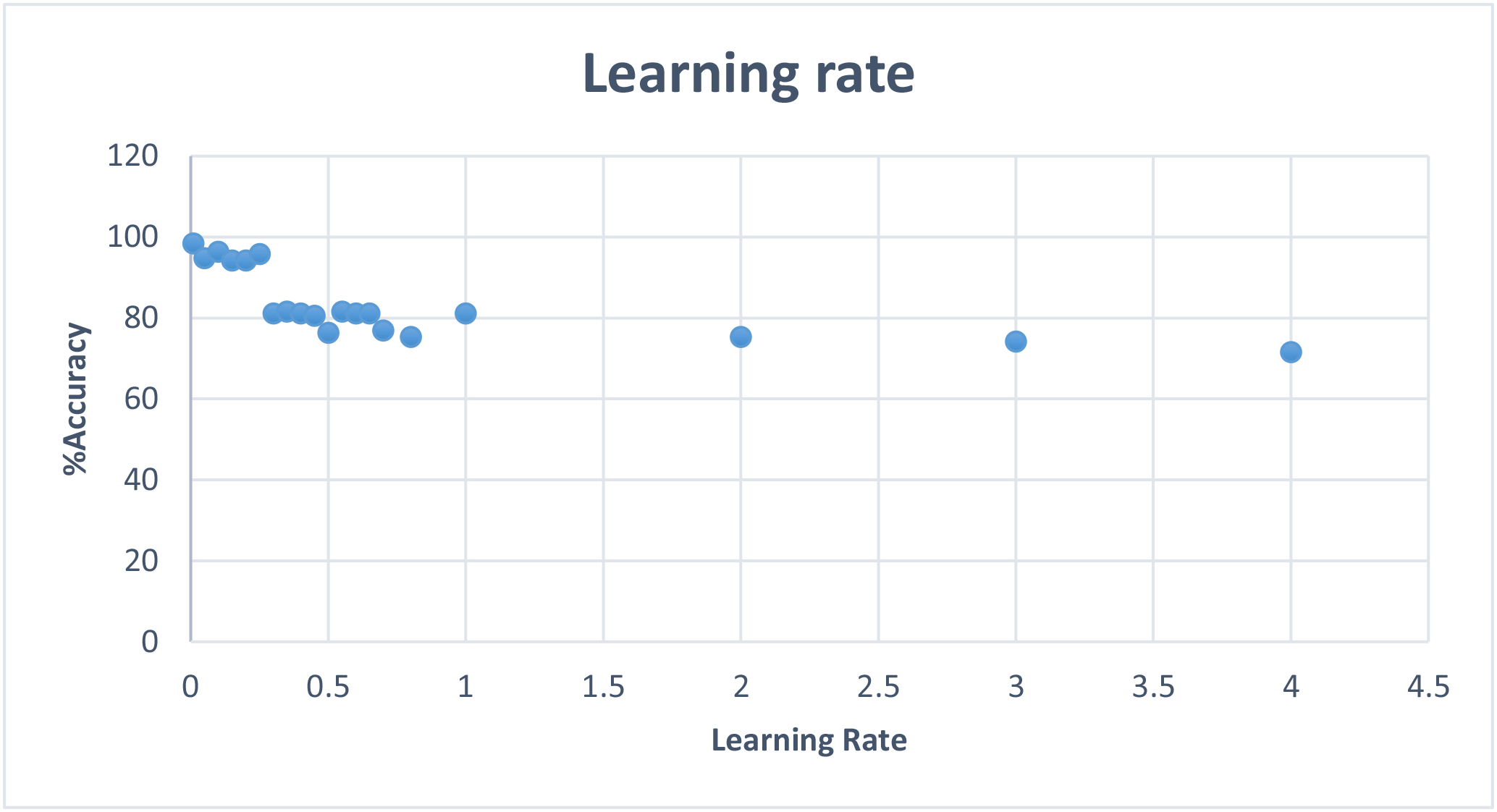

I observed that learning rate and max error rate were the most efficient at around 1% and increasing them had mostly negative effect on the effectiveness.
| Time | Efficiency in % | Normalised |
|---|---|---|
| 15.487, | 82.63157894736842 | yes |
| 15.009 | 98.42105263157895 | No |
Normalisation vs. no normalisation
| Time | Efficiency in % | Scaled down |
|---|---|---|
| 15.652, | 74.21052631578947 | Yes |
| 15.705, | 96.3157894736842 | No |
Down scaling (factor of 10) vs. no down scaling
As mentioned above, experimenting with scaling or normalising the data did not yield a positive effect, on the contrary, it reduced the classifier’s ability to make accurate predictions.
| Time | Efficiency in % | Activation function |
|---|---|---|
| 16.638 | 95.78947368421052 | SIGMOID |
| 16.37 | 92.63157894736842 | GAUSSIAN |
| 14.283 | 50.0 | LINEAR |
| 13.596 | 17.20430107526882 | TANH |
| 14.101, | 50.0 | TRAPEZOID |
Different activation functions
Comparison between methods
The difference between genetic programming and neural networks (MLP) was very noticeable. The neural network produced a more generic solution, as less knowledge was provided to it. To contrast that with the GP, many variables had to be tuned specifically to achieve the optimal result using syntax functions that are relevant to the solution. Neural networks are also faster and more efficient, outperforming the GP implementation. The implementation of the neural network is also a bit harder to reason with and the library hides most of the complexity away from the developer.
On the other hand, the Genetic Programming algorithm seemed more simple and intuitive, the tunable parameters had a direct impact on the result and the classification representation is simple. The program is also less generic as it required special tuning to achieve good results. It also does not provide consistent accuracy, where as the Neural Network had much smaller margin of error.
General note
The sourcecode folder contains gp_shell and neural_shell bash scripts that will work only on linux.
To manually run the java executable, use the following commands
java -cp CSC3423.jar Biocomputing.Neural.Control dataset/Training.arff
dataset/Test.arff
java -cp CSC3423.jar Biocomputing.GP.GPControl dataset/Training.arff
dataset/Test.arff
Where CSC3423.jar is the executable jar file in the out folder and the
arff files are the Training and Testing data sets.
The Neural network accepts the following environmental variables:
ITERATIONS, GENERATIONS,ACTIVATION_FUNCTION,
HIDDENNODES,MAXERROR,LEARNINGRATE,NORMALISE,SCALE.
GPClassifier environmental varialbes: POPULATION_SIZE, GENERATIONS,
ELITES, CROSSOVER_PROB, MUTATION_PROB, REPRODUCTION_PROB and
TOURNAMENT_SIZE.
In windows, one can use the command set HIDDENNODES=5 followed by the
java run command to set these values.
References
-
Moreira, M. (1995). Neural Networks with Adaptive Learning Rate and Momentum Terms. Available at https://infoscience.epfl.ch/record/82307/files/95-04.pdf. Accessed on 12.12.2016
-
Kun-Hong L. et al. (2008). A genetic programming-based approach to the classification of multiclass microarray datasets. Available at http://bioinformatics.oxfordjournals.org/content/25/3/331.full. Accessed on 14.12.2016
-
McCaffrey J. (2014). How To Standardize Data for Neural Networks. Available at https://visualstudiomagazine.com/articles/2014/01/01/how-to-standardize-data-for-neural-networks.aspx. Accessed on 14.12.2016.
-
Jeroen Eggermont et al (2004). Genetic Programming for Data Classification: Partitioning the Search Space. Available at http://liacs.leidenuniv.nl/~kosterswa/SAC2003final.pdf. Accessed on 15.12.2016
-
Álvarez L. et al. (2009). Artificial neural networks applied to cancer detection in a breast screening programme. Available at http://ac.els-cdn.com/S0895717710001378/1-s2.0-S0895717710001378-main.pdf?_tid=c4a6e3c0-c2b3-11e6-9ddd-00000aab0f6b&acdnat=1481798983_5428dbaa26e2d27c669d7906a5c1a77a. Accessed on 15.12.2016
-
Bengio Y., LeCun Y. (2007) Scaling Learning Algorithms towards AI. Available at http://yann.lecun.com/exdb/publis/pdf/bengio-lecun-07.pdf (Accessed: 16 Nov 2016).
Credits
-
Genetic programming tree image taken from http://www.ra.cs.uni-tuebingen.de/software/JCell/images/docbook/gp.png
-
Multilayer perceptron with back-propagation image taken from https://www.codeproject.com/KB/dotnet/predictor/network.jpg
Neural networks Genetic Programming MLP Machine Learning epochx classification sigmoidal